|
Information indices
|   |
List of information indices calculated by DRAGON
Information indices calculated by DRAGON are molecular descriptors defined as total and information content of molecules. Different criteria are used for defining equivalence classes, i.e. equivalency of atoms in a molecule such as chemical identity, ways of bonding through space, molecular topology and symmetry.
The total information content of a system having n elements is defined by the following:
![]()
where G is the number of different equivalence classes and ng is the number of elements in the gth class. Each equivalence class is built by the definition of some relationships among the elements of the system. The logarithm is taken at base 2 for measuring the information content in bits.
The total information content represents the residual information contained in the system after G relationships are defined among the n elements.
The mean information content, also called Shannon's entropy [C. Shannon, W. Weaver, The Mathematical Theory of Communication, University of Illinois Press, Urbana (ILL), 1949], is defined as:
![]()
Information indices based on molecule elements can be considered a quantitative measure of the lack of structural homogeneity or the diversity of a molecule, in this way being related to symmetry associated with structure.
Several information indices are derived from a graph representation of a molecule and are based on partitioning graph elements or matrix elements in equivalence classes according to two basic criteria [D. Bonchev, O. Mekenyan, N. Trinajstic, J.Comput.Chem. 1981, 2, 127-148; D. Bonchev, Information Theoretic Indices for Characterization of Chemical Structures, Research Studies Press, Chichester (UK), 1983]:
The information index on molecular size (ISIZ) is calculated as the total information content on the atom number [S.H. Bertz, J.Am.Chem.Soc. 1981, 103, 3599-3601], defined as:
![]()
where nAT is the number of molecule atoms (hydrogen included).
The total information index on atomic composition (IAC) and mean information index on atomic composition (AAC) are calculated as the total and mean information content, respectively, equivalence relationships being based on chemical atom types [S.M. Dancoff, H. Quastler, Essays on the Use of Information Theory in Biology, University of Illinois, Urbana (ILL), 1953]. Note that even hydrogens are considered in deriving these descriptors.
The total information content on the distance equality (IDET) and mean information content on the distance equality (IDE) are based on the equality of topological distances in a H-depleted molecular graph [D. Bonchev, N. Trinajstic, Int.J.Quantum Chem.Quant.Chem.Symp. 1978, 12, 293-303], the topological distance between two atoms being the length of the shortest path connecting the two atoms.
The total information content on the distance magnitude (IDMT) and mean information content on the distance magnitude (IDM) are based on the distribution of topological distances according to their magnitude in a H-depleted molecular graph, the topological distance between two atoms being the length of the shortest path connecting the two atoms.
The mean information content on the distance degree equality (IDDE) is based on the partition of vertex distance degrees according to their equality, the vertex distance degree of an atom being the sum of topological distances from the considered atom to any other atom in the H-depleted molecular graph.
The mean information content on the distance degree magnitude (IDDM) is based on the partition of vertex distance degrees according to their magnitude, the vertex distance degree of an atom being the sum of topological distances from the considered atom to any other atom in the H-depleted molecular graph.
The mean information content on the vertex degree equality (IVDE) is based on the partition of vertices according to vertex degree equality, the vertex degree of an atom being the number of connected non-H atoms.
The mean information content on the vertex degree magnitude (IVDM) is based on the partition of vertices according to the vertex degree magnitude, the vertex degree of an atom being the number of connected non-H atoms. This index was proposed as a measure of molecular complexity [C. Raychaudhury, S.K. Ray, J.J. Ghosh, A.B. Roy, S.C. Basak, J.Comput.Chem. 1984, 5, 581-588].
The graph vertex complexity index (HVcpx) is derived from the distance matrix where topological distances between pairs of atoms are collected as follows [C. Raychaudhury, S.K. Ray, J.J. Ghosh, A.B. Roy, S.C. Basak, J.Comput.Chem. 1984, 5, 581-588]:
![]()
where gfi is the number of distances from the vertex vi equal to g, hi is the atom eccentricity (i.e. the maximum topological distance from the vertex vi) and nSK the number of non-H atoms.
The graph distance complexity index (HDcpx) is derived from the distance matrix where topological distances between pairs of atoms are collected as follows [C. Raychaudhury, S.K. Ray, J.J. Ghosh, A.B. Roy, S.C. Basak, J.Comput.Chem. 1984, 5, 581-588; G. Klopman, C. Raychaudhury, R.V. Henderson, Math.Comput.Modelling 1988, 11, 635-640]:
![]()
where si is the ith vertex distance degree (i.e. sum of topological distances from the considered atom to any other atom), W is the Wiener index, dij is the topological distance between atoms i and j and nSK the number of molecule non-hydrogen atoms. DRAGON applies a logarithmic transformation to this index due to the large values it tends to have.
The Balaban U, V, X, Y indices (Uindex, Vindex, Xindex, Yindex) are calculated by the same formula as the Balaban distance connectivity index J using atomic information indices in place of vertex distance degrees [A.T. Balaban, T.-S. Balaban, J.Math.Chem. 1991, 8, 383-397].
The U index is based on atomic information indices ui calculated for vertices of a H-depleted molecular graph as follows:
![]()
where si is the ith vertex distance degree (i.e. sum of topological distances from the considered atom to any other atom), dij is the topological distance between atoms i and j and nSK the number of non-H atoms.
The V index (Vindex) is based on atomic information indices vi calculated for vertices of a H-depleted molecular graph as follows:
![]()
where si is the ith vertex distance degree (i.e. sum of topological distances from the considered atom to any other atom), dij is the topological distance between atoms i and j and nSK the number of non-H atoms.
The Y index is based on atomic information indices yi calculated for vertices of a H-depleted molecular graph as follows:
![]()
where g runs over all of the different topological distances from the ith vertex, gfi is the number of distances from the ith vertex equal to g, and hi is the ith atom eccentricity (i.e. the maximum topological distance from the considered atom).
The X index is based on atomic information indices xi calculated for vertices of a H-depleted molecular graph as follows:
![]()
where si is the vertex distance degree (i.e. sum of topological distances from the considered atom to any other atom), g runs over all of the different topological distances from the ith vertex, gfi is the number of distances from the ith vertex equal to g, and hi is the ith atom eccentricity (i.e. the maximum topological distance from the considered atom).
Indices of neighbourhood symmetry (ICk, TICk, SICk, BICk, CICk) are topological information indices calculated for a H-included molecular graph and based on neighbour degrees and edge multiplicity [V.R. Magnuson, D.K. Harriss, S.C. Basak, Topological Indices Based on Neighborhood Symmetry: Chemical and Biological Applications in Studies in Physical and Theoretical Chemistry, R.B. King (Ed.), Elsevier, Amsterdam (The Netherlands), pp. 178-191, 1983]. They are calculated by partitioning graph vertices into equivalence classes; the topological equivalence of two vertices is that the corresponding neighbourhoods of the kth order are the same. The vertex neighbourhood can be thought of as an open sphere comprising all the vertices in the graph, such that their distance from the considered vertex is less than k. DRAGON calculates Indices of neighbourhood symmetry from 0 up to 5 order.
The neighbourhood Information Content (ICk) is calculated as the mean information content as follows:
![]()
where g runs over the equivalence classes, Ag is the cardinality of the gth equivalence class and nAT is the total number of atoms. This index represents a measure of structural complexity per vertex.
The neighbourhood Total Information Content (TICk) is calculated as nAT times ICk, nAT being the total number of molecule atoms. This descriptor represents a measure of the graph complexity.
The Structural Information Content (SICk) is calculated in a normalised form of the information content ICk to delete the influence of graph size:
![]()
where nAT is the total number of molecule atoms.
The Bonding Information Content (BICk) is calculated in a normalised form of the information content ICk taking into account the number of bonds and their multiplicity:
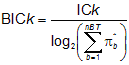
where nBT is the number of bonds and p* is the conventional bond order (1 for single, 2 for double, 3 for triple and 1.5 for aromatic bonds).
The Complementary Information Content (CICk) measures the deviation of the information content ICk from its maximum value, that corresponds to the vertex partition into equivalence classes containing one element each:
![]()
where nAT is the total number of molecule atoms.