|
WHIM descriptors
|   |
List of WHIM descriptors calculated by DRAGON
WHIM descriptors (Weighted Holistic Invariant Molecular descriptors) are geometrical descriptors based on statistical indices calculated on the projections of the atoms along principal axes [R.Todeschini, M.Lasagni, E.Marengo, J. Chemom. 1994, 8, 263-273; R.Todeschini, P.Gramatica, 3D QSAR in Drug Design - Vol. 2, H.Kubinyi, G.Folkers, Y.C.Martin (Eds.), Kluwer/ESCOM, Dordrecht (The Netherlands), 1998, 355-380].
WHIM descriptors are built in such a way as to capture relevant molecular 3D information regarding molecular size, shape, symmetry and atom distribution with respect to invariant reference frames. They are divided into two main classes: directional WHIM descriptors and global WHIM descriptors.
Directional WHIM descriptors are calculated as some univariate statistical indices on the projections of the atoms along each individual principal axis, while the global WHIMs are directly calculated as a combination of the former, thus simultaneously accounting for the variation of molecular properties along the three principal directions in the molecule. In this case, any information individually related to each principal axis disappears and the description is related only to a global view of the molecule.
Within the WHIM approach, a molecule is seen as a configuration of points (the atoms) in the three-dimensional space defined by the Cartesian axes (x,y,z). In order to obtain a unique reference frame, principal axes of the molecule are calculated. Then, projections of the atoms along each of the principal axes are performed and their dispersion and distribution around the geometric centre are evaluated.
Indeed, the algorithm consists of calculating the eigenvalues and eigenvectors of a weighted covariance matrix of the centred Cartesian coordinates of a molecule, obtained from different weighting schemes w for the atoms:
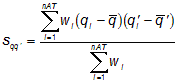
where sqq’ is the weighted covariance between the atomic coordinates q and q’ (q, q’ = x, y, z), nAT is the number of atoms, wi the atomic property, qi and q’i represent the coordinates of the ith atom, and ![]() the corresponding average value.
the corresponding average value.
A summary of WHIMs is shown in the table below.
|
Formula |
Symbol |
Molecular feature |
|
|
Lkw |
axial dimension |
|
|
Tw |
global dimension |
|
|
Aw |
global dimension |
|
|
Vw |
global dimension |
|
|
Pkw |
axial shape |
|
|
Kw |
shape |
|
|
Ekw |
axial density |
|
|
Dw |
global density |
|
|
Gkw |
axial symmetry |
|
|
Gw |
symmetry |
|
Table legend: l refers to eigenvalues of the weighted covariance matrix; t refers to atomic coordinates with respect to the principal axes; nAT is the number of molecule atoms; ns is the number of symmetric atoms along a principal axis and na the number of unsymmetric atoms. |
||
Six different weighting schemes are adopted: the unweighted case (u), atomic mass (m), the van der Waals volume (v), the Sanderson atomic electronegativity (e), the atomic polarizability (p) and the electrotopological state indices of Kier and Hall (s).
WHIM descriptors are invariant to translation due to the centering of the atomic coordinates and invariant to rotation due to the uniqueness of the principal axes, thus resulting free from prior alignment of molecules.
A fundamental role in the WHIM descriptor calculation is played by the eigenvalues l1, l2 and l3 of the weighted covariance matrix of the molecule atomic coordinates. Each eigenvalue represents a dispersion measure (i.e., the weighted variance) of the projected atoms along the considered principal axis, thus accounting for the molecular size along that principal direction. Relationships among the eigenvalues are used to describe the molecular shape. For example, for an ideal straight molecule both l2 and l3 are equal to zero and the global shape Kw is equal to 1 (maximum value); for an ideal spherical molecule all three eigenvalues are equal to 1/3 and Kw is 0.
Exploiting the new coordinates tk of the atoms along the principal axes, the atom distribution and density around the molecule centre are evaluated by an inverse function of the kurtosis k (h = 1/k ). Low values of the kurtosis are obtained when the atom projections assume opposite values with respect to the centre. When an increasing number of atom projections are within the extreme projections along a principal axis, the kurtosis value increases (i.e., kurtosis equal to 1.8 for a uniform distribution of points, to 3.0 for a normal distribution). When the kurtosis value tends to infinity the corresponding h value tends to zero.
In an analogous way, from the analysis of the new coordinates tk of the atoms, molecular symmetry is evaluated on the basis of the number ns of symmetric atoms with respect to the molecule centre, i.e., atoms with opposite coordinates along the considered axis, and the number na of unsymmetric atoms.