|
GETAWAY descriptors
|   |
List of GETAWAY descriptors calculated by DRAGON
The GETAWAY (GEometry, Topology, and Atom-Weights AssemblY) descriptors [V.Consonni, R.Todeschini, M.Pavan, J. Chem. Inf. Comput. Sci. 2002, 42, 682-692; V.Consonni, R.Todeschini, M.Pavan, P.Gramatica, J. Chem. Inf. Comput. Sci. 2002, 42, 693-705] have recently been proposed as chemical structure descriptors derived from a new representation of molecular structure, the Molecular Influence Matrix (MIM), denoted by H and defined as the following:
![]()
where M is the molecular matrix consisting of the centred Cartesian coordinates x, y, z of the molecule atoms (hydrogens included) in a chosen conformation, and the superscript T refers to the transposed matrix. Atomic coordinates are assumed to be calculated with respect to the geometrical centre of the molecule in order to obtain translation invariance. The molecular information matrix is a symmetric matrix and shows rotational invariance with respect to the molecule coordinates, thus resulting independent of molecule alignment.
The diagonal elements hii of the molecular influence matrix, called leverages, range from 0 to 1 and encode atomic information related to the "influence" of each molecule atom in determining the whole shape of the molecule; in effect, mantle atoms always have higher hii values than atoms near the molecule centre. Moreover, the magnitude of the maximum leverage in a molecule depends on the size and shape of the molecule. As derived from the geometry of the molecule, leverage values are effectively sensitive to significant conformational changes and to the bond lengths that account for atom types and bond multiplicity.
Each off-diagonal element hij represents the degree of accessibility of the jth atom to interactions with the ith atom, or, in other words, the attitude of the two considered atoms to interact with each other. A negative sign for the off-diagonal elements means that the two atoms occupy opposite molecular regions with respect to the center, hence the degree of their mutual accessibility should be low.
The influence/distance matrix R has been derived from the molecular influence matrix H as the following:
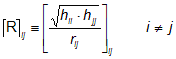
where hii and hjj are the leverages of the two considered atoms, and rij is their interatomic distance. The diagonal elements of the matrix R are zero. The squared root product of the leverages of two atoms is divided by their interatomic distance in order to make less significant contributions from pairs of atoms far apart, according to the basic idea that interactions between atoms in the molecule decreases as their distance increases.
The first set of the GETAWAY descriptors is shown in the table below. These descriptors have been derived by applying some traditional matrix operators and concepts of information theory both to the molecular influence matrix H and the influence/distance matrix R. Most of these descriptors are simply calculated only by the leverages used as the atomic weightings.
|
Formula |
Name |
|
|
geometric mean on the leverage magnitude |
|
|
total information content on the leverage equality |
|
|
standardized information content on the leverage equality |
|
|
mean information content on the leverage magnitude |
|
|
R matrix average row sum |
|
|
Randic-type R matrix connectivity |
|
|
first eigenvalue of the R matrix |
|
Table legend: nAT is the number of molecule atoms; nSK is the number of non-hydrogen atoms; Ng is the number of atoms with the same leverage value; G the number of equivalence classes; D = 1, 2 or 3 (1 for linear, 2 for planar and 3 for non-planar molecules); nBT is the number of molecule bonds. |
|
The geometric mean on the leverage magnitude (HGM) has been proposed to catch some information related to molecular shape. In effect, it has been found that in an isomeric series of hydrocarbons, the HGM increases from linear to more branched molecules; it is also inversely related to molecular size, decreasing as the number of atoms in the molecule increases.
The total and standardized information content on the leverage equality (ITH, ISH) mainly encode information on molecular symmetry; if all the atoms have different leverage values, i. e., the molecule does not show any element of symmetry, ITH = nSK*log(nSK) and ISH = 1; otherwise, if all the atoms have equal leverage values (a perfectly symmetric theoretical case), ITH = 0 and ISH = 0. The total information content on the leverage equality ITH is more discriminating than ISH for its dependence on molecular size, and thus it could be thought of as a measure of molecular complexity.
The mean information content on the leverage magnitude (HIC) seems to catch more information related to molecular complexity than the total and standardized information content on the leverage equality. Differently from ITH and ISH, HIC can, for example, recognise the different substituents in a series of monosubstituted benzenes. It is also sensitive to the presence of multiple bonds.
Both R matrix average row sum (RARS) and Randic-type R matrix connectivity (RCON) are based on the row sums of the influence/distance matrix since these encode some useful information that could be related to the presence of significant substituents or fragments in the molecule. In effect, it has been observed that larger row sums correspond to terminal atoms that are located very next to other terminal atoms such as those in substituents on a parent structure. Moreover, the RCON index is very sensitive to the molecular size as well as to conformational changes and cyclicity.
The first eigenvalue of the R matrix (REIG) has been defined on the analogy of the Lovasz-Pelikan index (descriptor LP1) that is an index of molecular branching calculated as the first eigenvalue of the adjacency matrix.
RARS and REIG indices are closely related; their values decrease as the molecular size increases and seem to be a little more sensitive to molecular branching than to cyclicity and conformational changes.
The other set of GETAWAY descriptors, shown in the table below, is based on the spatial autocorrelation formulas, weighting the molecule atoms by physico-chemical propertieswtogether with 3D information encoded by the elements of the molecular influence matrix H and influence/distance matrix R.
|
Formula |
Name |
|
|
leverage-weighted autocorrelation of lag k |
|
|
leverage-weighted total autocorrelation index |
|
|
H autocorrelation of lag k |
|
|
H total index |
|
|
R autocorrelation of lag k |
|
|
R total index |
|
|
R maximal autocorrelation of lag k |
|
|
R maximal index |
|
Table legend: nAT is the number of molecule atoms; dij is the topological distance between atoms i and j; wi is a physico-chemical atomic weight; d is the topological diameter; d(k; dij) is a Dirac-delta function (d=1 if dij = k, zero otherwise); d(k; dij; hij) is another Dirac-delta function (d = 1 if dij = k and hij>0, zero otherwise). |
|
The atomic properties w used for GETAWAY descriptor calculation are atomic mass (m), atomic polarizability (p), atomic electronegativity (e), van der Waals atomic volume (v), plus the unit weight (u).
HATS, H, R and maximal R indices are molecular descriptors for structure-property correlations, but they can also be used as molecular profiles suitable for similarity/diversity analysis studies. These descriptors, as based on spatial autocorrelation, encode information on structural fragments and therefore seem to be particularly suitable for describing differences in congeneric series of molecules. Differently from the Moreau-Broto autocorrelations, GETAWAYs are geometrical descriptors encoding information on the effective position of substituents and fragments in the molecular space. Moreover, they are independent of molecule alignment and, at some extent, account also for information on molecular size and shape as well as for specific atomic properties.