|
Eigenvalue-based indices
|   |
List of eigenvalue-based indices calculated by DRAGON
These molecular descriptors are calculated as a selected eigenvalue or function of the eigenvalues of a square matrix representing a H-depleted molecular graph.
The Lovasz-Pelikan index (LP1), also called leading eigenvalue, is the largest eigenvalue of the adjacency matrix [L. Lovasz, J. Pelikan, Period.Math.Hung. 1973, 3, 175-182].
Leading eigenvalues from weighted distance matrices Eig1w, w being an atomic property, are calculated as the largest eigenvalue of 5 weighted distance matrices accounting contemporarily for the presence of heteroatoms and multiple bonds in the molecule, defined as:
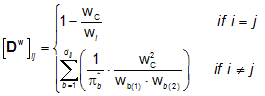
where wC is a property of the carbon atom, wi the property of the ith atom, p* is the conventional bond order (i.e. 1 for simple bond, 2 for double bond, 3 for triple bond and 1.5 for aromatic bond), the sum runs over all bonds involved in the shortest path between vertices i and j, dij being the topological distance (i.e. the length of the shortest path), and the subscripts b(1) and b(2) represent the two vertices incident to the considered b bond.
Atomic properties used in DRAGON to weight the distance matrix are: atomic number (Z), atomic mass (m), atomic van der Waals volume (v), atomic Sanderson electronegativity (e), and atomic polarizability (p).
The matrix weighted by atomic numbers Z is usually known as the Barysz distance matrix [M. Barysz, G. Jashari, R.S. Lall, A.K. Srivastava, N. Trinajstic, On the Distance Matrix of Molecules Containing Heteroatoms in Chemical Applications of Topology and Graph Theory, R.B. King (Ed.), Elsevier, Amsterdam (The Netherlands), pp. 222-230, 1983].
Eigenvalue sums from weighted distance matrices SEigw, w being an atomic property, are molecular descriptors calculated as the sum of all the eigenvalues of a weighted distance matrix.
Absolute eigenvalue sums from weighted distance matrices AEigw, w being an atomic property, are molecular descriptors calculated as the sum of the absolute values of all the eigenvalues of a weighted distance matrix.
VEM1 indices, M being a square matrix representing a H-depleted molecular graph, are molecular descriptors calculated as the sum of the coefficients of the eigenvector associated with the lowest (largest negative) eigenvalue of the matrix M [A.T. Balaban, D. Ciubotariu, M. Medeleanu, J.Chem.Inf.Comput.Sci. 1991, 31, 517-523]. VEM2 indices are the corresponding average values obtained by dividing each VEM1 index by the number of non-hydrogen atoms.
VRM1 indices, M being a square matrix representing a H-depleted molecular graph, are molecular descriptors calculated by a modified Randic-type formula using the coefficients of the eigenvector associated with the lowest (largest negative) eigenvalue of the matrix M in place of the vertex degrees [A.T. Balaban, D. Ciubotariu, M. Medeleanu, J.Chem.Inf.Comput.Sci. 1991, 31, 517-523].

The Randic-type formula has been modified in order to avoid too large values for big and symmetryc molecules for which some eigenvector coefficients tend towards zero.
VRM2 indices are the corresponding average values obtained by dividing each VRM1 index by the number of non-hydrogen atoms.
These molecular descriptors are based on the coefficients of the eigenvector associated with the lowest eigenvalue as they provide discrimination among molecule atoms; lower values correspond to atoms of lower connectivity, farther from the centre or from a vertex of high connectivity.
Molecular matrices M calculated by DRAGON with the corresponding VEM1, VEM2, VRM1 and VRM2 indices are: adjacency matrix A, distance matrix D, distance matrix weighted by atomic numbers Z, distance matrix weighted by atomic masses m, distance matrix weighted by atomic van der Waals volumes v, distance matrix weighted by atomic Sanderson electronegativities e and distance matrix weighted by atomic polarizabilities p.