|
Randic molecular profiles
|   |
List of Randic molecular profiles calculated by DRAGON
Molecular profiles are sequences of molecular descriptors proposed by Randic [M. Randic, New J.Chem. 1995, 19, 781-791; M. Randic, J.Chem.Inf.Comput.Sci. 1995, 35, 373-382; M. Randic, M. Razinger, J.Chem.Inf.Comput.Sci. 1995, 35, 594-606] and derived from the interatomic geometric distances of a molecule.
DRAGON provides two molecular profiles. One is much more related to the global molecular 3D structure:
(DP01, DP02, DP03, ... , DPk, ... , DP20)
and the other to the molecular shape:
(SP01, SP02, SP03, ..., SPk, ... , SP20)
Each descriptor DPk in the DP profile is calculated as:
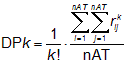
where rij is the geometric distance between atoms i and j, nAT the number of molecule atoms and k the descriptor order (k = 1, ... , 20). For large k values, DP values tend to zero, due to the effect of the factorial normalisation factor.
Each descriptor SPk of the shape profile is calculated in the same way as the DP descriptors, but taking into account only atoms on molecular periphery (i.e. atoms with H-depleted connectivity equal to 1 or 2).
Randic molecular profiles can be used in QSAR modelling, however they are particularly suitable to molecule similarity/diversity analysis since each profile well characterise a molecule.
The average shape profile index of order 2 (SHP2) is the shape profile index of order 2 (SP02) divided by the number of atoms with H-depleted connectivity equal to 1 or 2.